Section: New Results
Medical Image Analysis
Segmentation and Anatomical Variability of the Cochlea from Medical Images
Participants : Thomas Demarcy [Correspondant] , Hervé Delingette, Charles Raffaelli [CHU, Nice] , Clair Vandersteen [IUFC, Nice] , Dan Gnansia [Oticon Medical] , Nicholas Ayache.
This work is supported by the National Association for Research in Technology (ANRT) through the CIFRE Grant 2013-1165 and Oticon Medical (Vallauris). This work is a collaboration with the Department of Ear Nose Throat Surgery (IUFC, Nice) and the Nice University Hospital (CHU).
image segmentation ; surgery planning ; shape modelling ; anatomical variability ; cochlear implant ; temporal bone
-
We introduced an automated and reproducible framework for cochlear shape analysis [13].
-
We introduced a new cochlear segmentation method within a generative probabilistic Bayesian framework for CT images [6].
-
We studied the shape variability with a large database of CT images (N = 987) and quantified the bilateral symmetry in cochlear anatomy.
-
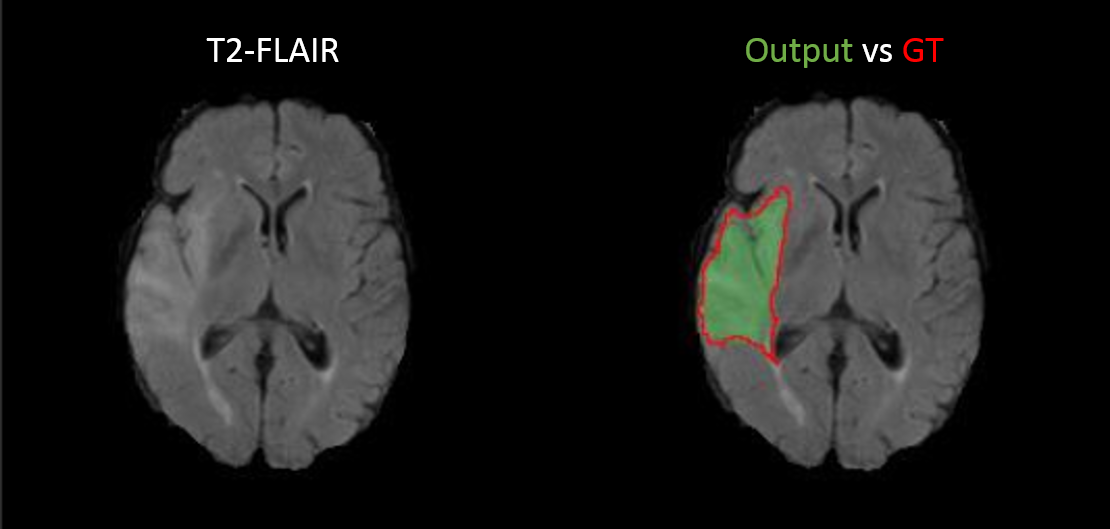
We provided a proof of concept for the estimation of postoperative cochlear implant electrode-array position from clinical CT (Fig. 1).
|
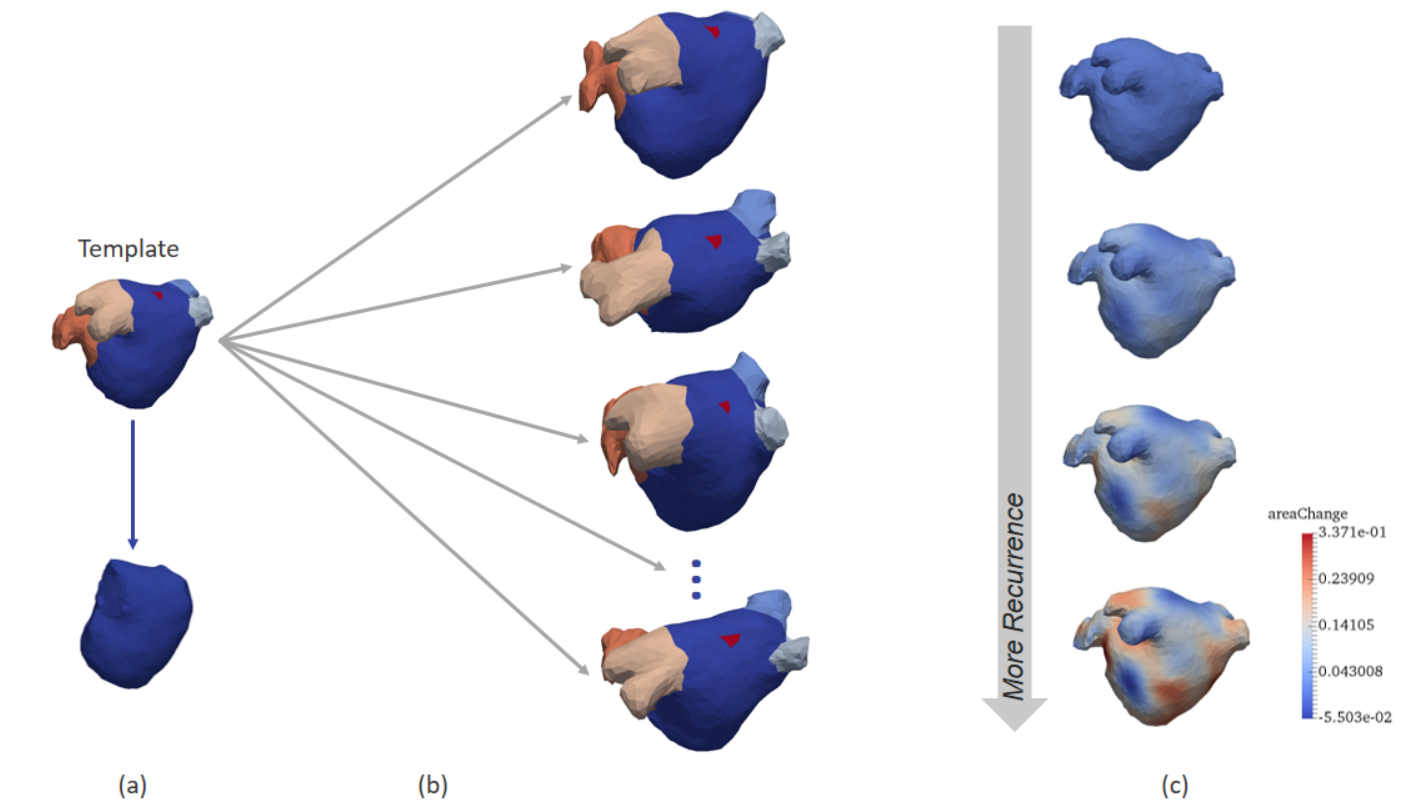
Prediction of Post-Ablation Outcome in Atrial Fibrillation Using Shape Parameterization and Partial Least Squares Regression
Participants : Shuman Jia [Correspondent] , Claudia Camaioni, Marc Michel Rohe, Pierre Jaïs, Xavier Pennec, Hubert Cochet, Maxime Sermesant.
The authors acknowledge the partial funding by the Agence Nationale de la Recherche (ANR)/ERA CoSysMedSysAFib and ANR MIGAT projects.
We proposed an application of diffeomorphometry and partial least squares regression to address the problem of post-ablation outcome in atrial fibrillation.
As illustrated in Fig. 2, we computed a template of left atrial shape in control group and then established point-to-point correspondence between patient-specific shapes and the template. The diffeomorphic deformations are encoded and applied in partial least squares regression to predict ablation success, which outperformed the left atrial volume index.
|
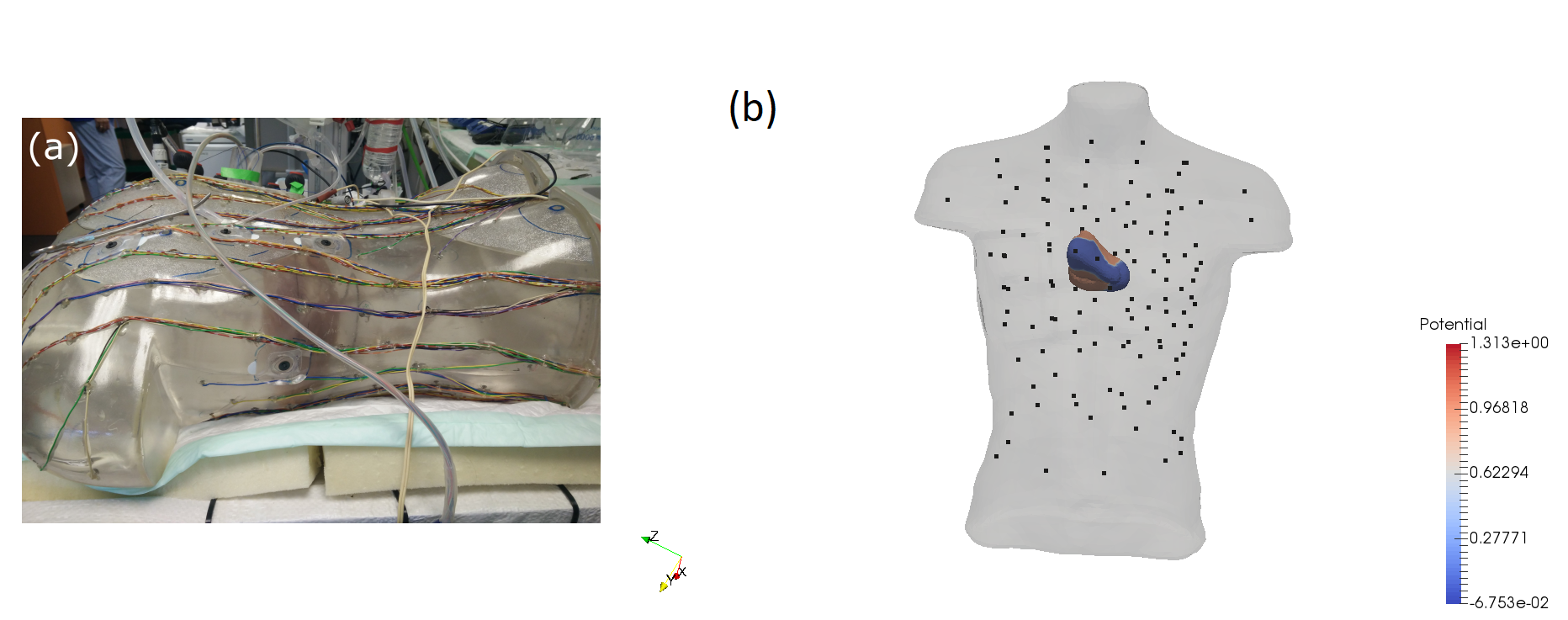
Cardiac Imaging and Machine Learning for Electrostructural Tomography
Participants : Tania Marina Bacoyannis [Correspondent] , Hubert Cochet [IHU Liryc, Bordeaux] , Maxime Sermesant.
This work is funded within the ERC Project ECSTATIC from the IHU Liryc, in Bordeaux.
Machine Learning, Cardiac modeling, Personalised simulation, Inverse problem of ECG, Electrical simulation, Inverse problem.
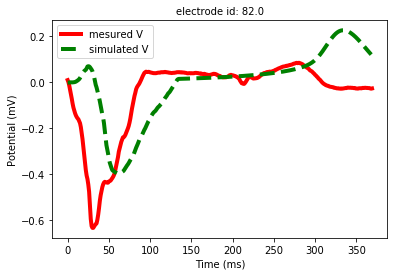
By using non-invasive electrical data (Body Surface Potential Mapping), we aim to develop a machine learning approach that can improve electrophysiological cardiac modeling in order to improve diagnosis and predict the response to therapy. This project involves measured and simulated data. For example, we processed experimental data provided by the IHU Liryc, gathered during an experiment on an healthy pig's heart (Figure 3). The simulated potentials appeared to be close to the measurements (Figure 4). The short-term goal is to reconstruct semi-automatically the simulated personalized activation maps.
|
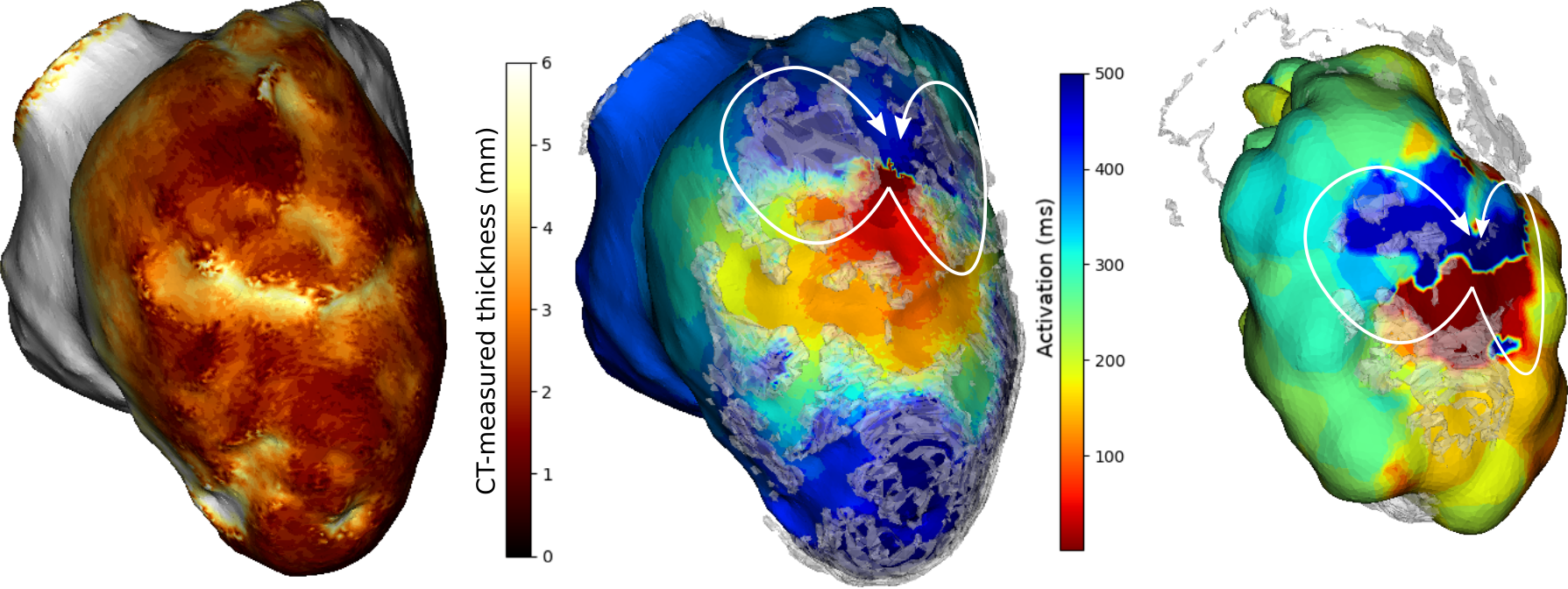
VT-Scan: image based modelling of cardiac electrophysiology to guide catheter radiofrequency ablation of re-entrant ventricular tachycardia
Participants : Nicolas Cedilnik [Correspondent] , Maxime Sermesant, Hubert Cochet, Pierre Jaïs, Frédéric Sacher.
This work was funded by IHU Liryc, Bordeaux.
cardiac electrophysiology modelling, cardiac imaging, ventricular tachycardia, catheter ablation, arrhythmia
-
We used cardiac CT images to estimate infarct scar density and location using an automated thickness computation.
-
A wavefront propagation speed was derived from this thickness in order to parametrize an Eikonal model of cardiac electrophysiology (see 5).
-
We were able to match our simulations to recorded ventricular tachycardia patterns obtained during catheter ablation procedures, on 10 different ventricular tachycardias.
-
This work was presented at the Functionnal Imaging and Modelling of the Heart conference in Toronto[34].
|
Deep Learning for Tumor Segmentation
Participants : Pawel Mlynarski [Correspondent] , Nicholas Ayache, Hervé Delingette, Antonio Criminisi [MSR] .
This work is funded by Inria-Microsoft joint center and is done in cooperation with Microsoft Research in Cambridge.
deep learning, semi-supervised learning, segmentation, MRI, tumors
|
-
We designed an algorithm for semi-supervised learning of neural nets for segmentation of tumors. The proposed system produces accurate binary segmentations (Figure 6) on unseen images with a limited number of ground truth segmentations used during the training phase.
-
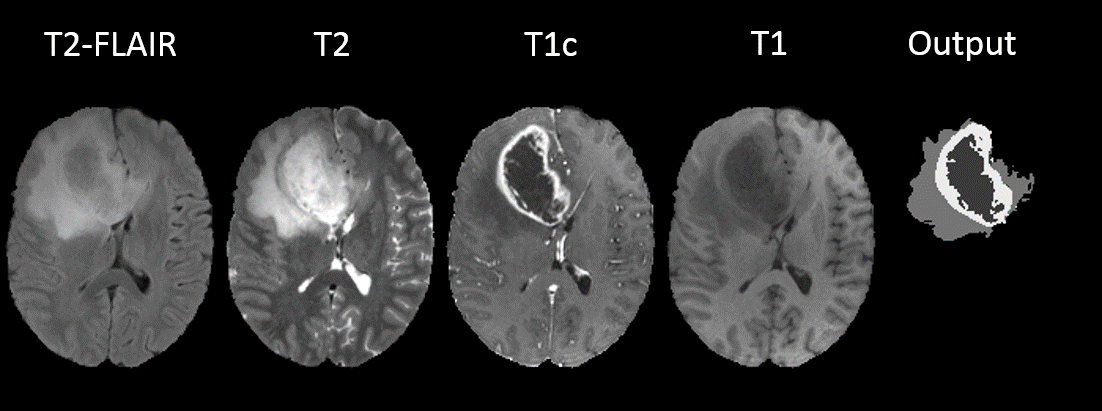
We proposed an efficient system based on Convolutional Neural Networks for multi-class segmentation of tumors in multimodal MR images (Figure 7). In particular, we proposed a new approach for treating different MR sequences and we introduced a new approach for ensembling 2D and 3D networks. We evaluated our method on the public benchmark of BRATS 2017 challenge and we obtained a top-3 performance among 60 participating teams.
Learning Brain Alterations in Multiple Sclerosis from Multimodal Neuroimaging Data
Participants : Wen Wei [Correspondent] , Nicholas Ayache [Inria] , Olivier Colliot [ARAMIS] .
Multiple Sclerosis, MRI, PET
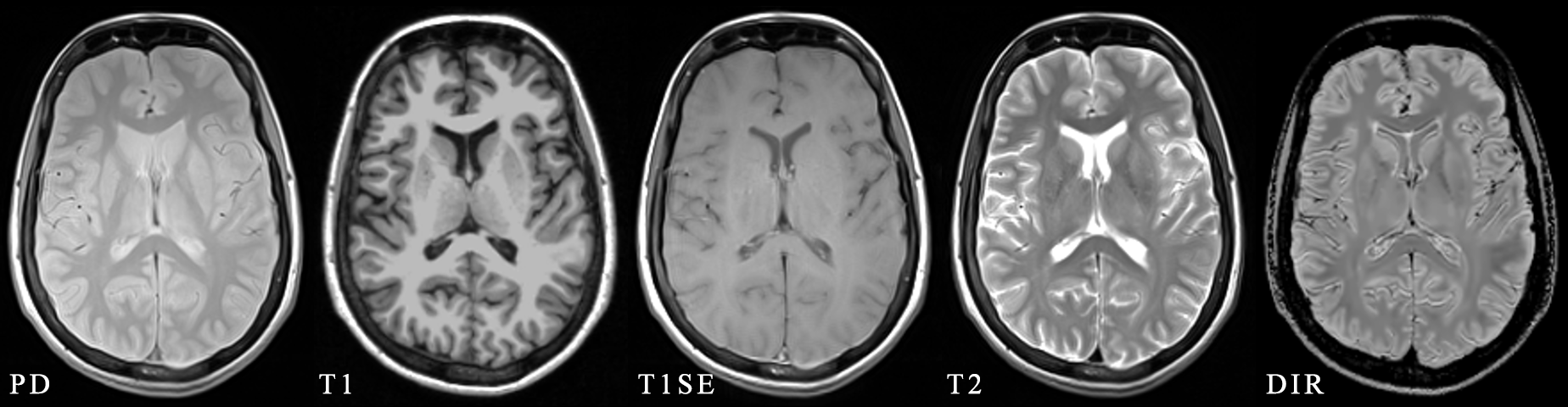
Multiple sclerosis (MS) is a demyelinating and inflammatory disease of the central nervous system . The goal of this topic is to develop a machine learning approach that can predict different types of PET-derived brain alterations using multiple local and regional MRI measures for MS patients. Figure 8 shows an example of multiple MRI pulse sequences for MS studies.
|
Robust and 3D-Consistent Cardiac Segmentation by Deep Learning
Participants : Qiao Zheng [Correspondent] , Hervé Delingette [Inria] , Nicolas Duchateau [Université Claude Bernard Lyon 1] , Nicholas Ayache [Inria] .
Cardiac Segmentation, Deep Learning, MRI, Robustness, Consistency
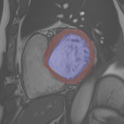
We propose a method based on deep learning to perform cardiac segmentation on short axis MRI image stacks. An example of segmentation is presented in Figure 9. The method is trained on a large database and then tested on other state-of-the-art cohorts. Results comparable or even better than the state-of-the-art in terms of distance measures are achieved. They prove the contribution of our method to enhance spatial consistency, and its generalization ability to unseen cases even from other databases.
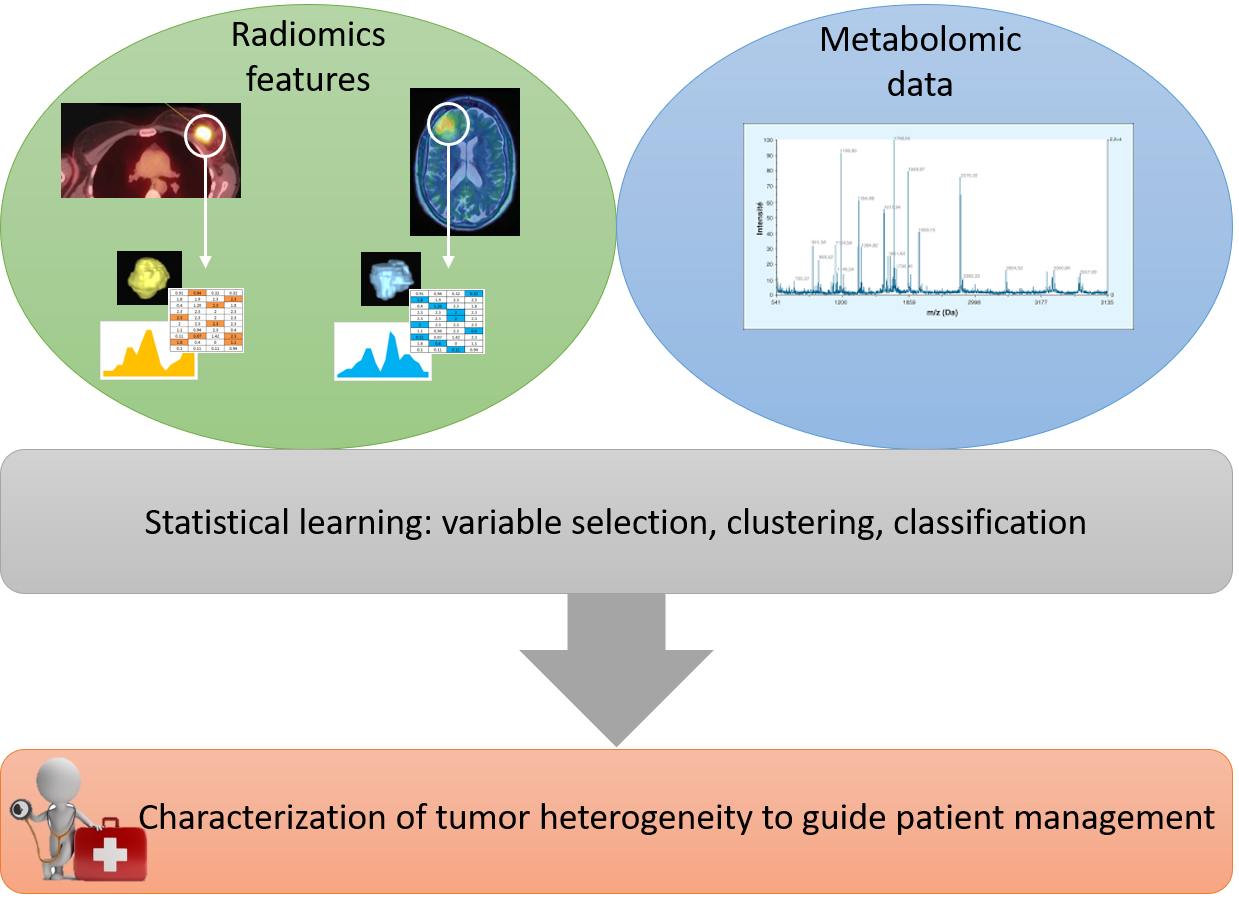
Joint analysis of radiomic and metabolomic features to improve diagnosis and therapy in oncology
Participants : Fanny Orlhac [Correspondent] , Charles Bouveyron, Hervé Delingette, Nicholas Ayache, Olivier Humbert [CAL] , Jacques Darcourt [CAL] , Thierry Pourcher [CEA] , Fanny Vandenbos [CHU Nice] .
Inria postdoctoral fellowship for 16 months
Radiomics, Metabolomics, Statistical learning
This work is done in collaboration with the Centre Antoine Lacassagne and the TIRO team (Transporter in Imagery and Radiotherapy for Oncology, CEA-UNS) located in Nice.
-
The project consists to jointly analyze histogram, shape and textural features extracted from medical images (radiomics) and metabolomic data in oncology (see Figure 10).
-
The goal is to better characterize tumor heterogeneity from both data sources in order to provide a personalized patient management.
-
The work focuses on two pathologies: breast cancer and glioblastoma.
Heart & Brain: discovering the link between cardiovascular pathologies and neurodegeneration through biophysical and statistical models of cardiac and brain images.
Participants : Jaume Banús Cobo [Correspondent] , Maxime Sermesant, Marco Lorenzi.
Uinversité Côte d'Azur (UCA)
Lumped models - Medical Imaging - Biophysical simulation - Machine learning
The project aims at developing a computational model of the relationship between cardiac function and brain damage from large‐scale clinical databases of multi‐modal and multi‐organ medical images. We will use advanced statistical learning tools for discovering relevant imaging features related to cardiac dysfunction and brain damage from large datasets of medical images and clinical information; these measurements will be combined within a unified mechanistic framework to understand and validate the relationship between cardiac function, vascular pathology and brain damage. The goal is to provide an unprecedented instrument for the in‐vivo assessment of latent neurodegenerative conditions in the general population, and will be validated with respect to established indices of cognitive decline and to specific sub‐population for which the ground truth is known.
|
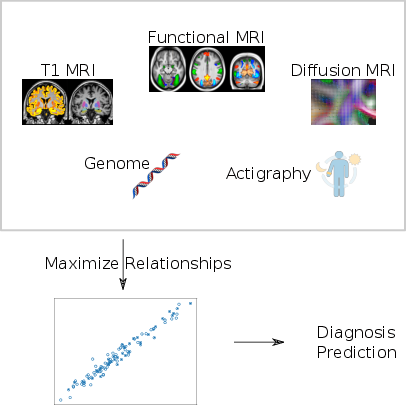
Statistical learning on large databases of heterogeneous imaging, cognitive and behavioural data
Participants : Luigi Antelmi [Correspondant] , Marco Lorenzi, Nicholas Ayache, Valeria Manera, Philippe Robert.
statistical learning, neuroimaging, big data, multimodal
The aim of our work is to develop scalable learning models for the joint analysis of heterogeneous biomedical data. The project will be applied to the investigation of neurological disorders from collections of brain imaging, body sensors, biological and clinical data available in current large-scale health databases. The resulting methodological framework will be tested on the UK Biobank, as well as on pathology-specific clinical data, as provided by the ADNI (http://adni.loni.usc.edu/), or INSIGHT (http://alzheimer-recherche.org/9248/etude-insight/) initiatives.
From the methodological perspective, the project will focus on the development of computationally efficient formulations of probabilistic latent variable models. These approaches will highlight meaningful relationship among biomarkers that will be used to develop optimal strategies for disease quantification and prediction (Fig. 12).
The research is within the MNC3 initiative (Médecine Numérique: Cerveau, Cognition, Comportement) funded by Université Côte d'Azur (UCA), and will be performed in collaboration with the Institut Claude Pompidou (CHU of Nice).
|
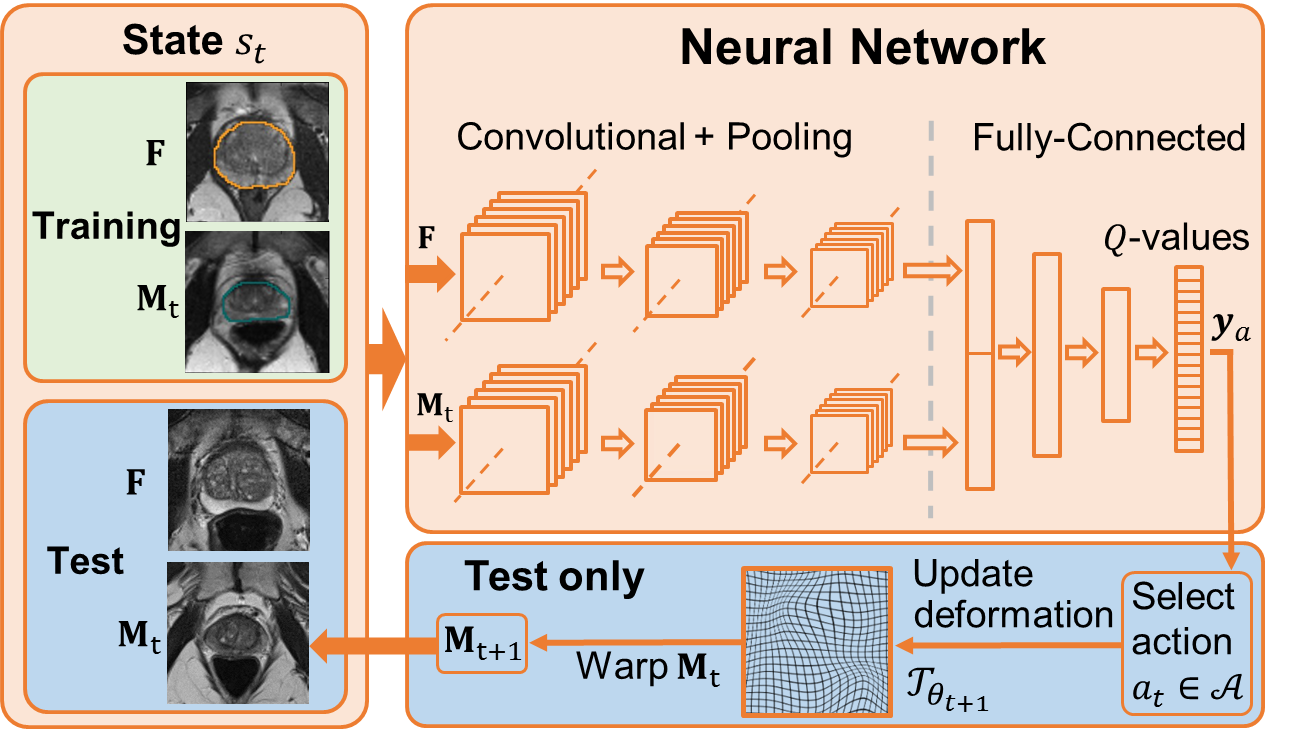
Robust non-rigid registration through agent-based action learning
Participants : Julian Krebs [Correspondent] , Hervé Delingette, Tommaso Mansi [Siemens [Siemens Healthineers, Medical Imaging Technologies] , Nicholas Ayache.
This PhD is carried out between the Asclepios research group, Inria Sophia Antipolis and Medical Imaging Technologies, Siemens Healthineers, Princeton, New Jersey, USA.
Deformable Registration, Deep Learning, Reinforcement Learning
We developed a deep learning-based approach for organ-specific deformable registration in 3-D [39] by:
-
reformulating deformable registration as an agent-based learning problem (Fig. 13)
-
using a low-parametric parametric statistical deformation model
-
applying a novel ground truth generator which allows generating millions of synthetically deformed training samples requiring only a few real deformation estimations
Improved performance has been demonstrated with respect to traditional algorithms.